Online Publications

Google Scholar Publication List
View all publications
Pubmed Publication List
View all publications2025 Journal Articles
Coherent Control over Nuclear Hyperpolarization Using an Optically Initializable Chromophore-Radical System
Chromophore radicals (CR) are emerging as important components for molecular quantum information science (QIS), especially in the context of quantum sensing. Here, we demonstrate that the optically hyperpolarized electrons in a 1,6,7,12-tetrakis(4-tert-butylphenoxy)-perylene-3,4,9,10-bis(dicarboximide) (tpPDI) covalently linked to a partially deuterated 1,3-bis(diphenylene)-d16-2-phenylallyl radical (BDPA-d16) can be coherently manipulated via pulsed dynamic nuclear polarization (DNP) methods to transfer polarization to nuclear spins and back. Under light illumination at 85 K, electron hyperpolarization in BDPA is enhanced 2.1- to 2.4-fold over thermal polarization and lasts for more than 100 ms. By applying nuclear orientation via electron spin-locking (NOVEL) DNP, this optically amplified electron hyperpolarization was successfully transferred to a 1H nuclear spin within the CR system and efficiently returned to the electron spin for readout via reverse-NOVEL. The NOVEL transfer efficiency of 65% amounts to a 688-fold nuclear spin hyperpolarization of the target nuclear spin, considering the 2.1-fold electron spin hyperpolarization. This reversible coherent manipulation of hyperpolarization transfer highlights the utility of CR systems to initialize and read out nuclear spin states in a disordered matrix at moderate cryogenic temperatures. Coupled with CRs’ environmental compatibility, tunability, and precise state initialization, these results highlight the promising role of nuclear spins in CRs for QIS applications, including quantum sensing and memory.
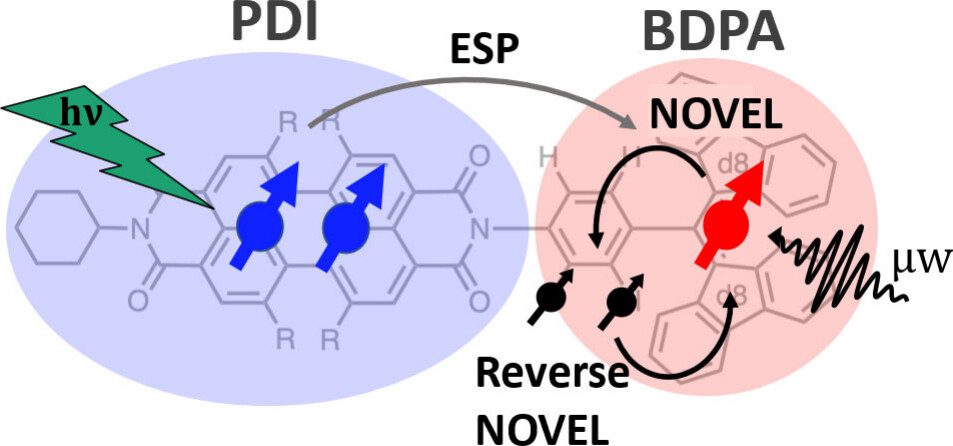
Coherent Control over Nuclear Hyperpolarization Using an Optically Initializable Chromophore-Radical System
Chromophore radicals (CR) are emerging as important components for molecular quantum information science (QIS), especially in the context of quantum sensing. Here, we demonstrate that the optically hyperpolarized electrons in a 1,6,7,12-tetrakis(4-tert-butylphenoxy)-perylene-3,4,9,10-bis(dicarboximide) (tpPDI) covalently linked to a partially deuterated 1,3-bis(diphenylene)-d16-2-phenylallyl radical (BDPA-d16) can be coherently manipulated via pulsed dynamic nuclear polarization (DNP) methods to transfer polarization to nuclear spins and back. Under light illumination at 85 K, electron hyperpolarization in BDPA is enhanced 2.1- to 2.4-fold over thermal polarization and lasts for more than 100 ms. By applying nuclear orientation via electron spin-locking (NOVEL) DNP, this optically amplified electron hyperpolarization was successfully transferred to a 1H nuclear spin within the CR system and efficiently returned to the electron spin for readout via reverse-NOVEL. The NOVEL transfer efficiency of 65% amounts to a 688-fold nuclear spin hyperpolarization of the target nuclear spin, considering the 2.1-fold electron spin hyperpolarization. This reversible coherent manipulation of hyperpolarization transfer highlights the utility of CR systems to initialize and read out nuclear spin states in a disordered matrix at moderate cryogenic temperatures. Coupled with CRs’ environmental compatibility, tunability, and precise state initialization, these results highlight the promising role of nuclear spins in CRs for QIS applications, including quantum sensing and memory.

Structure-specific Mini-Prion Model for Alzheimer’s Disease Tau Fibrils
A critical discovery of the past decade is that tau protein fibrils adopt disease-specific hallmark structures in each tauopathy. The faithful generation of synthetic fibrils adopting hallmark structures that can serve as targets for developing diagnostic and/or therapeutic strategies remains a grand challenge. We report on a rational design of synthetic fibrils built of a short peptide that adopts a critical structural motif in tauopathy fibrils found in Alzheimer’s Disease (AD) and Chronic Traumatic Encephalopathy (CTE). They serve as minimal prions with exquisite seeding competency, in vitro and in tau biosensor cells, for recruiting tau constructs ten times larger its size en route to AD or CTE fibril structures. We demonstrate that the generation of AD and CTE-like fibril structures is dramatically catalyzed in the presence of mini-AD prions and further influenced by salt composition in solution. Double Electron-Electron Resonance studies confirmed the preservation of AD-like folds across multi-generational seeding. Fibrils formed with the full AD/CTE-like core show strong seeding competency, with their templating effect dominating over the choice of salt composition that tunes the initial selection of AD- and CTE-like fibril populations. The mini-AD prions serve as a potent catalyst with templating capabilities that offer a novel strategy to design pathological tau fibril models.
Structure-specific Mini-Prion Model for Alzheimer’s Disease Tau Fibrils
A critical discovery of the past decade is that tau protein fibrils adopt disease-specific hallmark structures in each tauopathy. The faithful generation of synthetic fibrils adopting hallmark structures that can serve as targets for developing diagnostic and/or therapeutic strategies remains a grand challenge. We report on a rational design of synthetic fibrils built of a short peptide that adopts a critical structural motif in tauopathy fibrils found in Alzheimer’s Disease (AD) and Chronic Traumatic Encephalopathy (CTE). They serve as minimal prions with exquisite seeding competency, in vitro and in tau biosensor cells, for recruiting tau constructs ten times larger its size en route to AD or CTE fibril structures. We demonstrate that the generation of AD and CTE-like fibril structures is dramatically catalyzed in the presence of mini-AD prions and further influenced by salt composition in solution. Double Electron-Electron Resonance studies confirmed the preservation of AD-like folds across multi-generational seeding. Fibrils formed with the full AD/CTE-like core show strong seeding competency, with their templating effect dominating over the choice of salt composition that tunes the initial selection of AD- and CTE-like fibril populations. The mini-AD prions serve as a potent catalyst with templating capabilities that offer a novel strategy to design pathological tau fibril models.
Hydraulic Activation of the AsLOV2 photoreceptor
How proteins transduce environmental signals into mechanical motion remains a central question in biology. This study tests the hypothesis that blue light activation of AsLOV2 gives rise to concerted water movement that induce protein conformational extensions. Using electron and nuclear magnetic resonance spectroscopy, along with atomistic molecular dynamics simulations at high pressure, we find that activation, whether initiated by blue light or high pressure, is accompanied by selective expulsion of low-entropy, tetrahedrally coordinated “wrap” water from hydrophobic regions of the protein. These findings suggest that interfacial water serves as functional constituents to help reshape the protein’s free energy landscape during activation. Our study highlights hydration water as an active medium with the capacity to drive long-range conformational changes underlying protein mechanics and offers a new conceptual understanding for engineering externally controllable protein actuators for biomedical studies to smart materials.
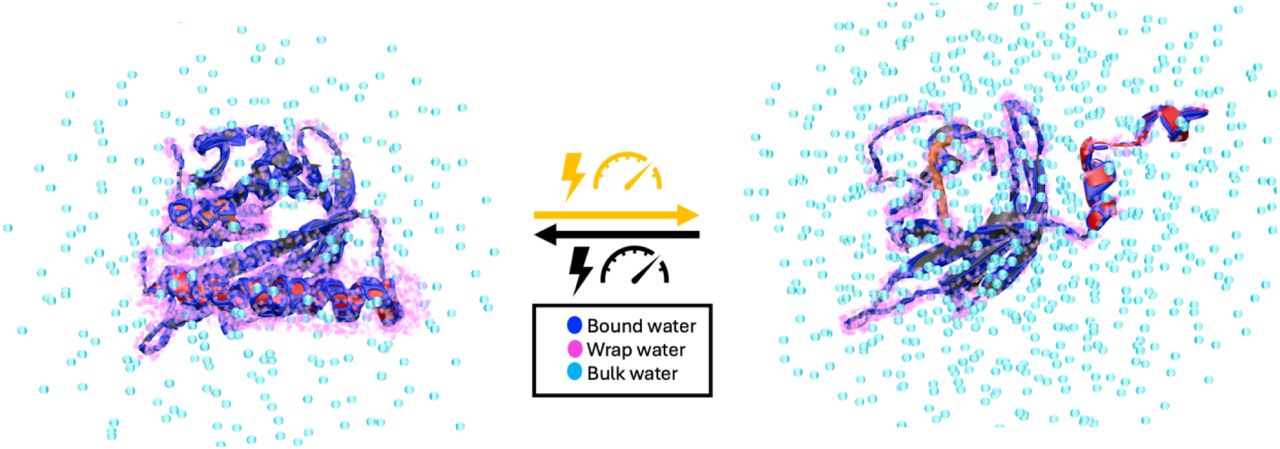
Hydraulic Activation of the AsLOV2 photoreceptor
How proteins transduce environmental signals into mechanical motion remains a central question in biology. This study tests the hypothesis that blue light activation of AsLOV2 gives rise to concerted water movement that induce protein conformational extensions. Using electron and nuclear magnetic resonance spectroscopy, along with atomistic molecular dynamics simulations at high pressure, we find that activation, whether initiated by blue light or high pressure, is accompanied by selective expulsion of low-entropy, tetrahedrally coordinated “wrap” water from hydrophobic regions of the protein. These findings suggest that interfacial water serves as functional constituents to help reshape the protein’s free energy landscape during activation. Our study highlights hydration water as an active medium with the capacity to drive long-range conformational changes underlying protein mechanics and offers a new conceptual understanding for engineering externally controllable protein actuators for biomedical studies to smart materials.

Water-directed pinning is key to tau prion formation
Featured in Northwestern Now, Phys.org, & Le Monde.
Tau forms fibrillar aggregates that are pathological hallmarks of a family of neurodegenerative diseases known as tauopathies. The synthetic replication of disease-specific fibril structures is a critical gap for developing diagnostic and therapeutic tools. This study debuts a strategy of identifying a critical and minimal folding motif in fibrils characteristic of tauopathies and generating seeding-competent fibrils from the isolated tau peptides. The 19-residue jR2R3 peptide (295 to 313) which spans the R2/R3 splice junction of tau, and includes the P301L mutation, is one such peptide that forms prion-competent fibrils. This tau fragment contains the hydrophobic VQIVYK hexapeptide that is part of the core of all known pathological tau fibril structures and an intramolecular counterstrand that stabilizes the strand–loop–strand (SLS) motif observed in 4R tauopathy fibrils. This study shows that P301L exhibits a duality of effects: it lowers the barrier for the peptide to adopt aggregation-prone conformations and enhances the local structuring of water around the mutation site to facilitate site-directed pinning and dewetting around sites 300-301 to achieve in-register stacking of tau to cross β-sheets. We solved a 3 Å cryo-EM structure of jR2R3-P301L fibrils in which each protofilament layer contains two jR2R3-P301L copies, of which one adopts a SLS fold found in 4R tauopathies and the other wraps around the SLS fold to stabilize it, reminiscent of the three- and fourfold structures observed in 4R tauopathies. These jR2R3-P301L fibrils are competent to template full-length 4R tau in a prion-like manner.
Water-directed pinning is key to tau prion formation
Featured in Northwestern Now, Phys.org, & Le Monde.
Tau forms fibrillar aggregates that are pathological hallmarks of a family of neurodegenerative diseases known as tauopathies. The synthetic replication of disease-specific fibril structures is a critical gap for developing diagnostic and therapeutic tools. This study debuts a strategy of identifying a critical and minimal folding motif in fibrils characteristic of tauopathies and generating seeding-competent fibrils from the isolated tau peptides. The 19-residue jR2R3 peptide (295 to 313) which spans the R2/R3 splice junction of tau, and includes the P301L mutation, is one such peptide that forms prion-competent fibrils. This tau fragment contains the hydrophobic VQIVYK hexapeptide that is part of the core of all known pathological tau fibril structures and an intramolecular counterstrand that stabilizes the strand–loop–strand (SLS) motif observed in 4R tauopathy fibrils. This study shows that P301L exhibits a duality of effects: it lowers the barrier for the peptide to adopt aggregation-prone conformations and enhances the local structuring of water around the mutation site to facilitate site-directed pinning and dewetting around sites 300-301 to achieve in-register stacking of tau to cross β-sheets. We solved a 3 Å cryo-EM structure of jR2R3-P301L fibrils in which each protofilament layer contains two jR2R3-P301L copies, of which one adopts a SLS fold found in 4R tauopathies and the other wraps around the SLS fold to stabilize it, reminiscent of the three- and fourfold structures observed in 4R tauopathies. These jR2R3-P301L fibrils are competent to template full-length 4R tau in a prion-like manner.

Hydrolytically Stable Phosphonate‐Based Metal–Organic Frameworks for Harvesting Water from Low Humidity Air
Harvesting water from air offers a promising solution to the global water crisis. However, existing sorbents often struggle in arid climates due to limitations such as low sorption capacities, hydrolytic instability, slow mass transport, high desorption enthalpy, and costly operation. Phosphonate-based metal–organic frameworks (MOFs), known for their exceptional water stability, have not been extensively explored for water harvesting. This study systematically investigates the performance of STA-12 (M═Co, Ni, Mg) and STA-16 (M═Co, Ni), a series of stable phosphonate-based MOFs, as water sorbents. STA-12 MOFs demonstrate remarkable adsorption at ultra-low humidity (<10%), while STA-16(Co) exhibits a high water uptake capacity of 0.54 g g−1 at 10–50% relative humidity (RH) and 0.72 g g−1 at 34% RH. Molecular simulations and solid-state NMR identified liquid-like water, critical for harvesting applications, as the key contributor to the superior sorption performance of STA-16(Co). Scalable aqueous synthesis methods are developed, producing tens of grams of MOFs per batch without high-pressure equipment. A prototype device incorporating STA-12(Ni) demonstrated the feasibility of these materials for real-world water harvesting, showcasing their potential to address water scarcity in arid regions
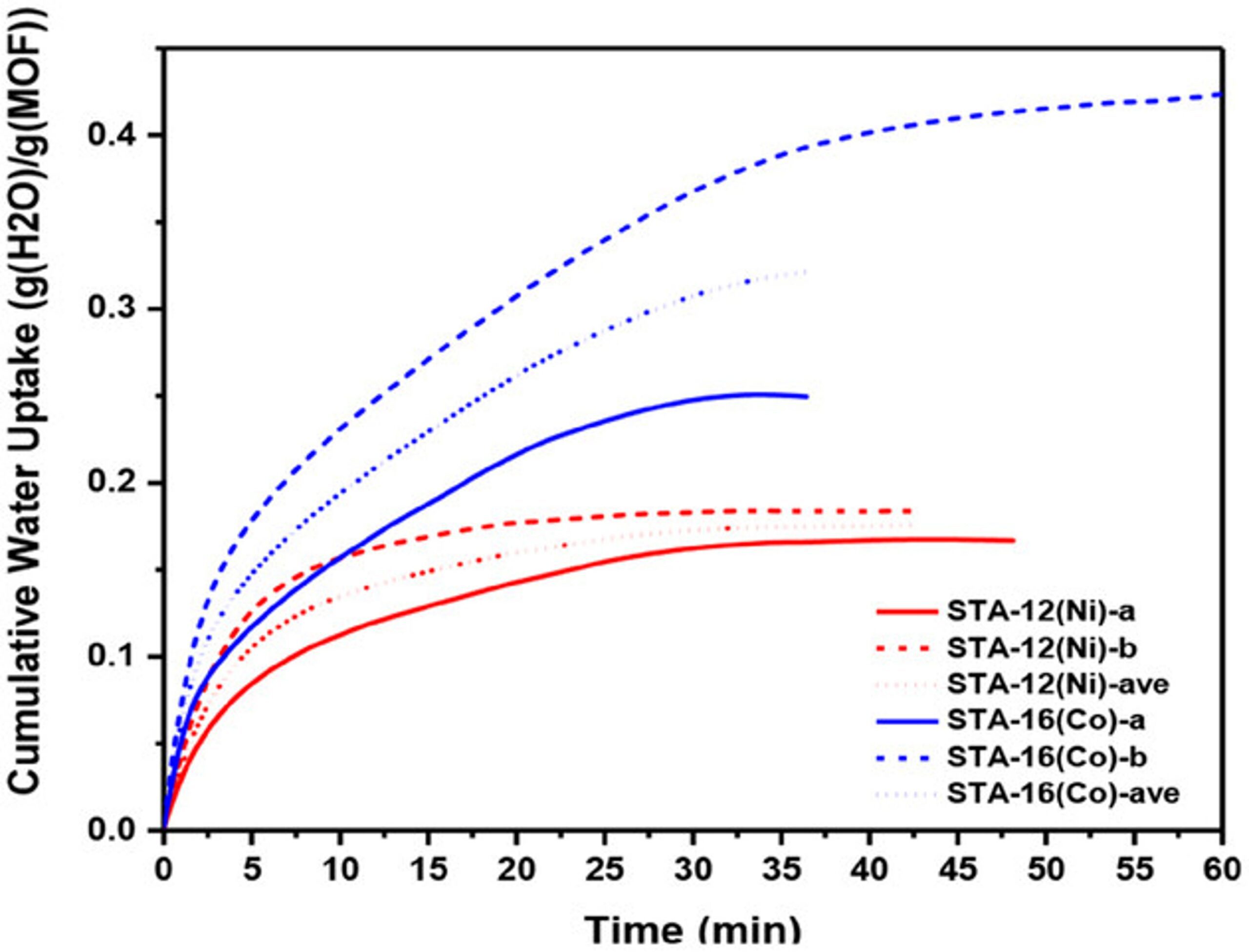
Hydrolytically Stable Phosphonate‐Based Metal–Organic Frameworks for Harvesting Water from Low Humidity Air
Harvesting water from air offers a promising solution to the global water crisis. However, existing sorbents often struggle in arid climates due to limitations such as low sorption capacities, hydrolytic instability, slow mass transport, high desorption enthalpy, and costly operation. Phosphonate-based metal–organic frameworks (MOFs), known for their exceptional water stability, have not been extensively explored for water harvesting. This study systematically investigates the performance of STA-12 (M═Co, Ni, Mg) and STA-16 (M═Co, Ni), a series of stable phosphonate-based MOFs, as water sorbents. STA-12 MOFs demonstrate remarkable adsorption at ultra-low humidity (<10%), while STA-16(Co) exhibits a high water uptake capacity of 0.54 g g−1 at 10–50% relative humidity (RH) and 0.72 g g−1 at 34% RH. Molecular simulations and solid-state NMR identified liquid-like water, critical for harvesting applications, as the key contributor to the superior sorption performance of STA-16(Co). Scalable aqueous synthesis methods are developed, producing tens of grams of MOFs per batch without high-pressure equipment. A prototype device incorporating STA-12(Ni) demonstrated the feasibility of these materials for real-world water harvesting, showcasing their potential to address water scarcity in arid regions

Passaging Human Tauopathy Patient Samples in Cells Generates Heterogeneous Fibrils with a Subpopulation Adopting Disease Folds
The discovery by cryo-electron microscopy (cryo-EM) that the neu-ropathological hallmarks of different tauopathies, including Alzheimer’s disease, corticobasal degeneration (CBD), and progressive supranuclear palsy (PSP), are caused by unique misfolded conformations of the protein tau is among the most profound developments in neurodegenerative disease research. To capitalize on these discoveries for therapeutic development, one must achieve in vitroreplication of tau fibrils that adopt the representative tauopathy disease folds, which represents a grand challenge for the field. A widely used approach has been seeded propagation using pathological tau fibrils derived from post-mortem patient samples in biosensor cells that expresses a fragment of the tau protein known as K18, or Tau4RD, containing the microtubule-binding repeat domain of tau as the substrate. The new insights from cryo-EM raised the question of whether the Tau4RD fragment is capable of adopting characteristic tau folds found in CBD and PSP patient fibrils, and whether cell-passaged and amplified tau fibrils can be used as seeds to achieve cell-free assembly of recombinant 4R tau into fibrils without the addition of cofactors. Using Double Electron Electron Resonance (DEER) spectroscopy, we discovered that cell-passaged pathological seeds generate heterogeneous fibrils that are, however, distinct between the CBD and PSP lysate-seeded fibrils, and vastly different from heparin-induced tau fibril structures. Moreover, the lysate-seeded fibrils contain a characteristic sub-population that resembles the disease fold corresponding to the respective starting patient sample. These findings indicate that templated propagation using CBD and PSP patient-derived fibrils is possible with a tau fragment that does not contain the entire pathological fibril core, but also that additional mechanisms must be tuned to converge on a homogeneous fibril population.

Passaging Human Tauopathy Patient Samples in Cells Generates Heterogeneous Fibrils with a Subpopulation Adopting Disease Folds
The discovery by cryo-electron microscopy (cryo-EM) that the neu-ropathological hallmarks of different tauopathies, including Alzheimer’s disease, corticobasal degeneration (CBD), and progressive supranuclear palsy (PSP), are caused by unique misfolded conformations of the protein tau is among the most profound developments in neurodegenerative disease research. To capitalize on these discoveries for therapeutic development, one must achieve in vitroreplication of tau fibrils that adopt the representative tauopathy disease folds, which represents a grand challenge for the field. A widely used approach has been seeded propagation using pathological tau fibrils derived from post-mortem patient samples in biosensor cells that expresses a fragment of the tau protein known as K18, or Tau4RD, containing the microtubule-binding repeat domain of tau as the substrate. The new insights from cryo-EM raised the question of whether the Tau4RD fragment is capable of adopting characteristic tau folds found in CBD and PSP patient fibrils, and whether cell-passaged and amplified tau fibrils can be used as seeds to achieve cell-free assembly of recombinant 4R tau into fibrils without the addition of cofactors. Using Double Electron Electron Resonance (DEER) spectroscopy, we discovered that cell-passaged pathological seeds generate heterogeneous fibrils that are, however, distinct between the CBD and PSP lysate-seeded fibrils, and vastly different from heparin-induced tau fibril structures. Moreover, the lysate-seeded fibrils contain a characteristic sub-population that resembles the disease fold corresponding to the respective starting patient sample. These findings indicate that templated propagation using CBD and PSP patient-derived fibrils is possible with a tau fragment that does not contain the entire pathological fibril core, but also that additional mechanisms must be tuned to converge on a homogeneous fibril population.

Localized Reconstruction of Multimodal Distance Distribution from DEER Data of Biopolymers
Pulsed Dipolar ESR Spectroscopy (PDS) is a uniquely powerful technique to characterize the structural property of intrinsically disordered proteins (IDPs) and polymers and the conformational evolution of IDPs and polymers, e.g. during assembly, by offering the probability distribution of segment end-to-end distances. However, it is challenging to determine distance distribution P(r) of IDPs by PDS because of the uncertain and broad shape information that is intrinsic to the distance distribution of IDPs. We demonstrate here that the Srivastava-Freed Singular Value Decomposition (SF-SVD) point-wise mathematical inversion method along with wavelet denoising (WavPDS) can aid in obtaining reliable shapes for the distance distribution, P(r), for IDPs. We show that broad regions of P(r) as well as mixed narrow and broad features within the captured distance distribution range can be effectively resolved and differentiated without a priori knowledge. The advantage of SF-SVD and WavPDS is that the methods are transparent, requiring no adjustable parameters, the processing of the magnitude for the probability distribution is performed separately for each distance increment, and the outcome of the analysis is independent of the user’s judgement. We demonstrate the performance and present the application of WavPDS and SF-SVD on model ruler molecules, model polyethylene glycol polymers with end-to-end spin labeling, and IDPs with pairwise labeling spanning different segments of the protein tau to generate the transparent solutions to the P(r)’s including their uncertainties and error analysis.

Localized Reconstruction of Multimodal Distance Distribution from DEER Data of Biopolymers
Pulsed Dipolar ESR Spectroscopy (PDS) is a uniquely powerful technique to characterize the structural property of intrinsically disordered proteins (IDPs) and polymers and the conformational evolution of IDPs and polymers, e.g. during assembly, by offering the probability distribution of segment end-to-end distances. However, it is challenging to determine distance distribution P(r) of IDPs by PDS because of the uncertain and broad shape information that is intrinsic to the distance distribution of IDPs. We demonstrate here that the Srivastava-Freed Singular Value Decomposition (SF-SVD) point-wise mathematical inversion method along with wavelet denoising (WavPDS) can aid in obtaining reliable shapes for the distance distribution, P(r), for IDPs. We show that broad regions of P(r) as well as mixed narrow and broad features within the captured distance distribution range can be effectively resolved and differentiated without a priori knowledge. The advantage of SF-SVD and WavPDS is that the methods are transparent, requiring no adjustable parameters, the processing of the magnitude for the probability distribution is performed separately for each distance increment, and the outcome of the analysis is independent of the user’s judgement. We demonstrate the performance and present the application of WavPDS and SF-SVD on model ruler molecules, model polyethylene glycol polymers with end-to-end spin labeling, and IDPs with pairwise labeling spanning different segments of the protein tau to generate the transparent solutions to the P(r)’s including their uncertainties and error analysis.

Phosphoryl group wires stabilize pathological tau fibrils as revealed by multiple quantum spin counting NMR
Hyperphosphorylation of the protein tau is one of the biomarkers of neurodegenerative diseases in the category of tauopathies. However, the molecular level, mechanistic, role of this common post- translational modification (PTM) in enhancing or reducing the aggregation propensity of tau is unclear, especially considering that combinatorial phosphorylation of multiple sites can have complex, non-additive, effects on tau protein aggregation. Since tau proteins stack in register and parallel to elongate into pathological fibrils, phosphoryl groups from adjacent tau strands with 4.8 Å separation must find an energetically favorable spatial arrangement. At first glance, this appears to be an unfavorable configuration due to the proximity of negative charges between phosphate groups from adjacent neighboring tau fibrils. However, this study tests a counterhypothesis that phosphoryl groups within the fibril core-forming segments favorably assemble into highly ordered, hydrogen-bonded, one-dimensionally extended wires under biologically relevant conditions. We selected two phosphorylation sites associated with neurodegeneration, serine 305 (S305p) and tyrosine 310 (Y310p), on a model tau peptide jR2R3-P301L (tau295-313) spanning the R2/R3 splice junction of tau, that readily aggregate into a fibril with characteristics of a seed-competent mini prion. Using multiple quantum spin counting (MQ-SC) by 31P solid-state NMR of phosphorylated jR2R3-P301L tau peptide fibrils, enhanced by dynamic nuclear polarization, we find that at least six phosphorous spins must neatly arrange in 1D within fibrils or in 2D within a protofibril to yield the experimentally observed MQ-coherence orders of four. We found that S305pstabilizes the tau fibrils and leads to more seeding-competent fibrils compared to jR2R3 P301L or Y310p. This study introduces a new concept that phosphorylation of residues within a core forming tau segment can mechanically facilitate fibril registry and stability due a hitherto unrecognized role of phosphoryl groups to form highly ordered, extended, 1D wires that stabilize pathological tau fibrils.
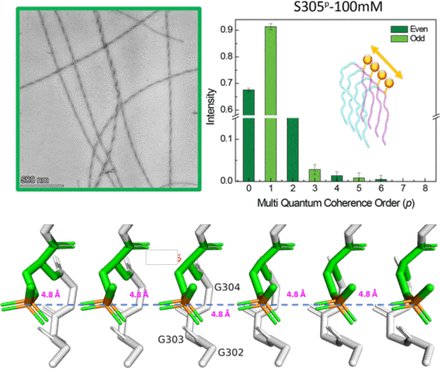
Phosphoryl group wires stabilize pathological tau fibrils as revealed by multiple quantum spin counting NMR
Hyperphosphorylation of the protein tau is one of the biomarkers of neurodegenerative diseases in the category of tauopathies. However, the molecular level, mechanistic, role of this common post- translational modification (PTM) in enhancing or reducing the aggregation propensity of tau is unclear, especially considering that combinatorial phosphorylation of multiple sites can have complex, non-additive, effects on tau protein aggregation. Since tau proteins stack in register and parallel to elongate into pathological fibrils, phosphoryl groups from adjacent tau strands with 4.8 Å separation must find an energetically favorable spatial arrangement. At first glance, this appears to be an unfavorable configuration due to the proximity of negative charges between phosphate groups from adjacent neighboring tau fibrils. However, this study tests a counterhypothesis that phosphoryl groups within the fibril core-forming segments favorably assemble into highly ordered, hydrogen-bonded, one-dimensionally extended wires under biologically relevant conditions. We selected two phosphorylation sites associated with neurodegeneration, serine 305 (S305p) and tyrosine 310 (Y310p), on a model tau peptide jR2R3-P301L (tau295-313) spanning the R2/R3 splice junction of tau, that readily aggregate into a fibril with characteristics of a seed-competent mini prion. Using multiple quantum spin counting (MQ-SC) by 31P solid-state NMR of phosphorylated jR2R3-P301L tau peptide fibrils, enhanced by dynamic nuclear polarization, we find that at least six phosphorous spins must neatly arrange in 1D within fibrils or in 2D within a protofibril to yield the experimentally observed MQ-coherence orders of four. We found that S305pstabilizes the tau fibrils and leads to more seeding-competent fibrils compared to jR2R3 P301L or Y310p. This study introduces a new concept that phosphorylation of residues within a core forming tau segment can mechanically facilitate fibril registry and stability due a hitherto unrecognized role of phosphoryl groups to form highly ordered, extended, 1D wires that stabilize pathological tau fibrils.

2024 Journal Articles
Field-domain rapid-scan EPR at 240 GHz for studies of protein functional dynamics at room temperature
We present field-domain rapid-scan (RS) electron paramagnetic resonance (EPR) at 8.6 T and 240 GHz. To enable this technique, we upgraded a homebuilt EPR spectrometer with an FPGA-enabled digitizer and real-time processing software. The software leverages the Hilbert transform to recover the in-phase (𝐼) and quadrature (𝑄) channels, and therefore the raw absorptive and dispersive signals, 𝜒′ and 𝜒′′, from their combined magnitude (√𝐼2+𝑄2). Averaging a magnitude is simpler than real-time coherent averaging and has the added benefit of permitting long-timescale signal averaging (up to at least 2.5 × 106 scans) because it eliminates the effects of sourcereceiver phase drift. Our rapid-scan (RS) EPR provides a signal-to-noise ratio that is approximately twice that of continuous wave (CW) EPR under the same experimental conditions, after scaling by the square root of acquisition time. We apply our RS EPR as an extension of the recently reported time-resolved Gd-Gd EPR (TiGGER) [Maity et al., 2023], which is able to monitor inter-residue distance changes during the photocycle of a photoresponsive protein through changes in the Gd-Gd dipolar couplings. RS, opposed to CW, returns field-swept spectra as a function of time with 10 ms time resolution, and thus, adds a second dimension to the static field transients recorded by TiGGER. We were able to use RS TiGGER to track time-dependent and temperature-dependent kinetics of AsLOV2, a light-activated phototropin domain found in oats. The results presented here combine the benefits of RS EPR with the improved spectral resolution and sensitivity of Gd chelates at high magnetic fields. In the future, field-domain RS EPR at high magnetic fields may enable studies of other real-time kinetic processes with time resolutions that are otherwise difficult to access in the solution state.
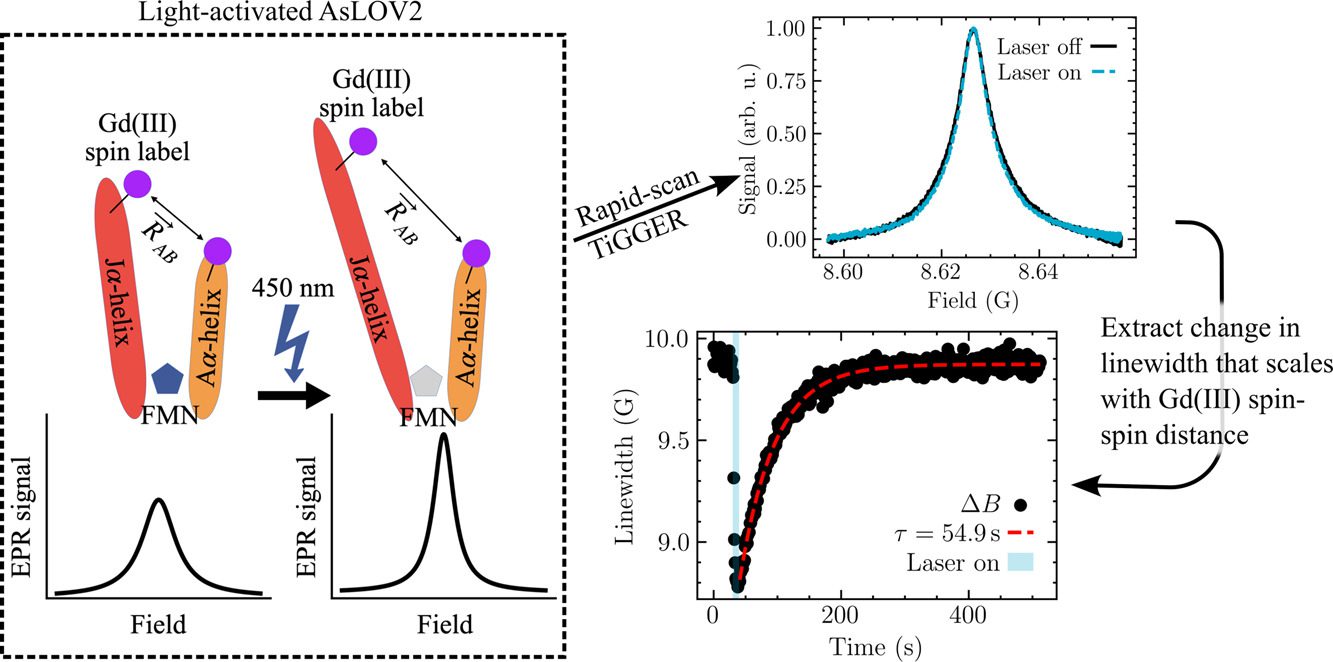
Field-domain rapid-scan EPR at 240 GHz for studies of protein functional dynamics at room temperature
We present field-domain rapid-scan (RS) electron paramagnetic resonance (EPR) at 8.6 T and 240 GHz. To enable this technique, we upgraded a homebuilt EPR spectrometer with an FPGA-enabled digitizer and real-time processing software. The software leverages the Hilbert transform to recover the in-phase (𝐼) and quadrature (𝑄) channels, and therefore the raw absorptive and dispersive signals, 𝜒′ and 𝜒′′, from their combined magnitude (√𝐼2+𝑄2). Averaging a magnitude is simpler than real-time coherent averaging and has the added benefit of permitting long-timescale signal averaging (up to at least 2.5 × 106 scans) because it eliminates the effects of sourcereceiver phase drift. Our rapid-scan (RS) EPR provides a signal-to-noise ratio that is approximately twice that of continuous wave (CW) EPR under the same experimental conditions, after scaling by the square root of acquisition time. We apply our RS EPR as an extension of the recently reported time-resolved Gd-Gd EPR (TiGGER) [Maity et al., 2023], which is able to monitor inter-residue distance changes during the photocycle of a photoresponsive protein through changes in the Gd-Gd dipolar couplings. RS, opposed to CW, returns field-swept spectra as a function of time with 10 ms time resolution, and thus, adds a second dimension to the static field transients recorded by TiGGER. We were able to use RS TiGGER to track time-dependent and temperature-dependent kinetics of AsLOV2, a light-activated phototropin domain found in oats. The results presented here combine the benefits of RS EPR with the improved spectral resolution and sensitivity of Gd chelates at high magnetic fields. In the future, field-domain RS EPR at high magnetic fields may enable studies of other real-time kinetic processes with time resolutions that are otherwise difficult to access in the solution state.

Salt-Screened Transition toward Bulk-Like Water Dynamics near Polymeric Zwitterions
Harvesting water from air offers a promising solution to the global water crisis. However, existing sorbents often struggle in arid climates due to limitations such as low sorption capacities, hydrolytic instability, slow mass transport, high desorption enthalpy, and costly operation. Phosphonate-based metal–organic frameworks (MOFs), known for their exceptional water stability, have not been extensively explored for water harvesting. This study systematically investigates the performance of STA-12 (M═Co, Ni, Mg) and STA-16 (M═Co, Ni), a series of stable phosphonate-based MOFs, as water sorbents. STA-12 MOFs demonstrate remarkable adsorption at ultra-low humidity (<10%), while STA-16(Co) exhibits a high water uptake capacity of 0.54 g g−1 at 10–50% relative humidity (RH) and 0.72 g g−1 at 34% RH. Molecular simulations and solid-state NMR identified liquid-like water, critical for harvesting applications, as the key contributor to the superior sorption performance of STA-16(Co). Scalable aqueous synthesis methods are developed, producing tens of grams of MOFs per batch without high-pressure equipment. A prototype device incorporating STA-12(Ni) demonstrated the feasibility of these materials for real-world water harvesting, showcasing their potential to address water scarcity in arid regions
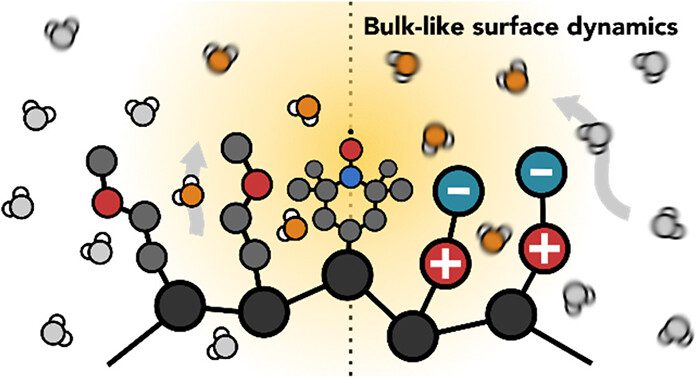
Salt-Screened Transition toward Bulk-Like Water Dynamics near Polymeric Zwitterions
Harvesting water from air offers a promising solution to the global water crisis. However, existing sorbents often struggle in arid climates due to limitations such as low sorption capacities, hydrolytic instability, slow mass transport, high desorption enthalpy, and costly operation. Phosphonate-based metal–organic frameworks (MOFs), known for their exceptional water stability, have not been extensively explored for water harvesting. This study systematically investigates the performance of STA-12 (M═Co, Ni, Mg) and STA-16 (M═Co, Ni), a series of stable phosphonate-based MOFs, as water sorbents. STA-12 MOFs demonstrate remarkable adsorption at ultra-low humidity (<10%), while STA-16(Co) exhibits a high water uptake capacity of 0.54 g g−1 at 10–50% relative humidity (RH) and 0.72 g g−1 at 34% RH. Molecular simulations and solid-state NMR identified liquid-like water, critical for harvesting applications, as the key contributor to the superior sorption performance of STA-16(Co). Scalable aqueous synthesis methods are developed, producing tens of grams of MOFs per batch without high-pressure equipment. A prototype device incorporating STA-12(Ni) demonstrated the feasibility of these materials for real-world water harvesting, showcasing their potential to address water scarcity in arid regions

Dynamic Nuclear Polarization Using Electron Spin Cluster
Dynamic nuclear polarization (DNP) utilizing narrow-line electron spin clusters (ESCs) to achieve nuclear spin resonance matching (ESC-DNP) by microwave irradiation is a promising way to achieve NMR signal enhancements with a wide design scope requiring low microwave power at high magnetic field. Here we present the design for a trityl-based tetra-radical (TetraTrityl) to achieve DNP for 1H NMR at 7 T, supported by experimental data and quantum mechanical simulations. A slow-relaxing (T1e ≈ 1 ms) 4-ESC is found to require at least two electron spin pairs at <8 Å e–e spin distance to yield 1H ESC-DNP enhancement, while squeezing the rest of the e–e spin distances to <12 Å results in optimal 1H ESC-DNP enhancements. Fast-relaxing ESCs (T1e ≈ 10 μs) are found to require a weakly coupled narrow-line radical (sensitizer) to extract polarization from the ESC. These results provide design principles for achieving a power-efficient DNP at high field via ESC-DNP.
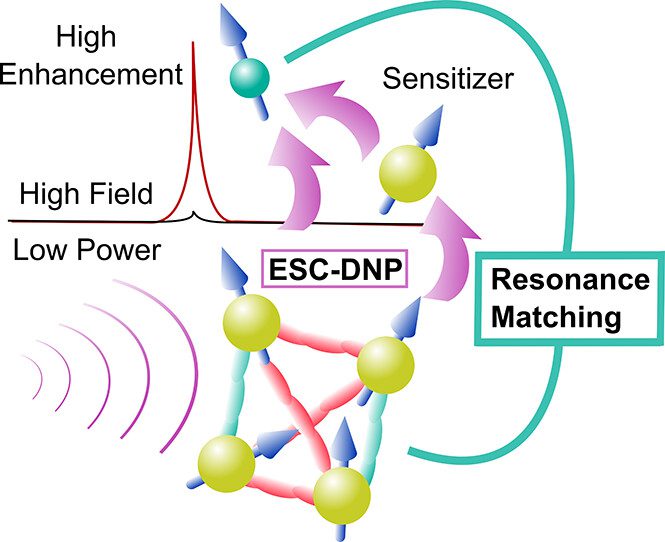
Dynamic Nuclear Polarization Using Electron Spin Cluster
Dynamic nuclear polarization (DNP) utilizing narrow-line electron spin clusters (ESCs) to achieve nuclear spin resonance matching (ESC-DNP) by microwave irradiation is a promising way to achieve NMR signal enhancements with a wide design scope requiring low microwave power at high magnetic field. Here we present the design for a trityl-based tetra-radical (TetraTrityl) to achieve DNP for 1H NMR at 7 T, supported by experimental data and quantum mechanical simulations. A slow-relaxing (T1e ≈ 1 ms) 4-ESC is found to require at least two electron spin pairs at <8 Å e–e spin distance to yield 1H ESC-DNP enhancement, while squeezing the rest of the e–e spin distances to <12 Å results in optimal 1H ESC-DNP enhancements. Fast-relaxing ESCs (T1e ≈ 10 μs) are found to require a weakly coupled narrow-line radical (sensitizer) to extract polarization from the ESC. These results provide design principles for achieving a power-efficient DNP at high field via ESC-DNP.

Dynamic Nuclear Polarization Enhanced Multiple Quantum Spin Counting of Molecular Assemblies in Vitrified Solutions
Crystallization pathways are essential to various industrial, geological, and biological processes. In nonclassical nucleation theory, prenucleation clusters (PNCs) form, aggregate, and crystallize to produce higher order assemblies. Microscopy and X-ray techniques have limited utility for PNC analysis due to the small size (0.5–3 nm) and time stability constraints. We present a new approach for analyzing PNC formation based on 31P nuclear magnetic resonance (NMR) spin counting of vitrified molecular assemblies. The use of glassing agents ensures that vitrification generates amorphous aqueous samples and offers conditions for performing dynamic nuclear polarization (DNP)-amplified NMR spectroscopy. We demonstrate that molecular adenosine triphosphate along with crystalline, amorphous, and clustered calcium phosphate materials formed via a nonclassical growth pathway can be differentiated from one another by the number of dipolar coupled 31P spins. We also present an innovative approach for examining spin counting data, demonstrating that a knowledge-based fitting of integer multiples of cosine wave functions, instead of the traditional Fourier transform, provides a more physically meaningful retrieval of the existing frequencies. This is the first report of multiquantum spin counting of assemblies formed in solution as captured under vitrified DNP conditions, which can be useful for future analysis of PNCs and other aqueous molecular clusters.
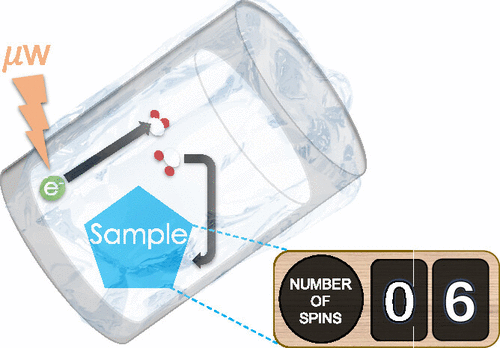
Dynamic Nuclear Polarization Enhanced Multiple Quantum Spin Counting of Molecular Assemblies in Vitrified Solutions
Crystallization pathways are essential to various industrial, geological, and biological processes. In nonclassical nucleation theory, prenucleation clusters (PNCs) form, aggregate, and crystallize to produce higher order assemblies. Microscopy and X-ray techniques have limited utility for PNC analysis due to the small size (0.5–3 nm) and time stability constraints. We present a new approach for analyzing PNC formation based on 31P nuclear magnetic resonance (NMR) spin counting of vitrified molecular assemblies. The use of glassing agents ensures that vitrification generates amorphous aqueous samples and offers conditions for performing dynamic nuclear polarization (DNP)-amplified NMR spectroscopy. We demonstrate that molecular adenosine triphosphate along with crystalline, amorphous, and clustered calcium phosphate materials formed via a nonclassical growth pathway can be differentiated from one another by the number of dipolar coupled 31P spins. We also present an innovative approach for examining spin counting data, demonstrating that a knowledge-based fitting of integer multiples of cosine wave functions, instead of the traditional Fourier transform, provides a more physically meaningful retrieval of the existing frequencies. This is the first report of multiquantum spin counting of assemblies formed in solution as captured under vitrified DNP conditions, which can be useful for future analysis of PNCs and other aqueous molecular clusters.

Dipolar Order Induced Electron Spin Hyperpolarization
The structure of coupled electron spin systems is of fundamental interest to many applications, including dynamic nuclear polarization (DNP), enhanced nuclear magnetic resonance (NMR), the generation of electron spin qubits for quantum information science (QIS), and quantitative studies of paramagnetic systems by electron paramagnetic resonance (EPR). However, the characterization of electron spin coupling networks is nontrivial, especially at high magnetic fields. This study focuses on a system containing high concentrations of trityl radicals that give rise to a DNP enhancement profile of 1H NMR characteristic of the presence of electron spin clusters. When this system is subject to selective microwave saturation through pump–probe ELectron DOuble Resonance (ELDOR) experiments, electron spin hyperpolarization is observed. We show that the generation of an out-of-equilibrium longitudinal dipolar order is responsible for the transient hyperpolarization of electron spins. Notably, the coupled electron spin system needs to form an AX-like system (where the difference in the Zeeman interactions of two spins is larger than their coupling interaction) such that selective microwave irradiation can generate signatures of electron spin hyperpolarization. We show that the extent of dipolar order, as manifested in the extent of electron spin hyperpolarization generated, can be altered by tuning the pump or probe pulse length, or the interpulse delay in ELDOR experiments that change the efficiency to generate or readout longitudinal dipolar order. Pump–probe ELDOR with selective saturation is an effective means for characterizing coupled electron spins forming AX-type spin systems that are foundational for DNP and quantum sensing.
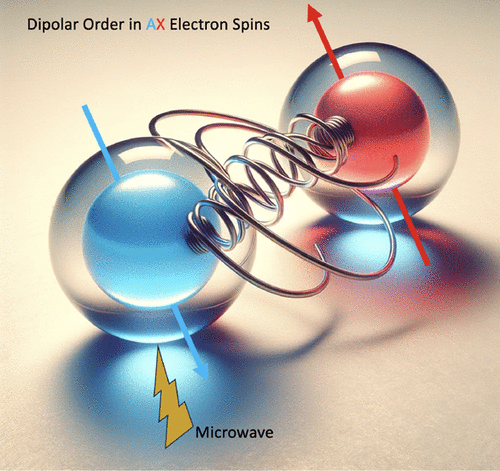
Dipolar Order Induced Electron Spin Hyperpolarization
The structure of coupled electron spin systems is of fundamental interest to many applications, including dynamic nuclear polarization (DNP), enhanced nuclear magnetic resonance (NMR), the generation of electron spin qubits for quantum information science (QIS), and quantitative studies of paramagnetic systems by electron paramagnetic resonance (EPR). However, the characterization of electron spin coupling networks is nontrivial, especially at high magnetic fields. This study focuses on a system containing high concentrations of trityl radicals that give rise to a DNP enhancement profile of 1H NMR characteristic of the presence of electron spin clusters. When this system is subject to selective microwave saturation through pump–probe ELectron DOuble Resonance (ELDOR) experiments, electron spin hyperpolarization is observed. We show that the generation of an out-of-equilibrium longitudinal dipolar order is responsible for the transient hyperpolarization of electron spins. Notably, the coupled electron spin system needs to form an AX-like system (where the difference in the Zeeman interactions of two spins is larger than their coupling interaction) such that selective microwave irradiation can generate signatures of electron spin hyperpolarization. We show that the extent of dipolar order, as manifested in the extent of electron spin hyperpolarization generated, can be altered by tuning the pump or probe pulse length, or the interpulse delay in ELDOR experiments that change the efficiency to generate or readout longitudinal dipolar order. Pump–probe ELDOR with selective saturation is an effective means for characterizing coupled electron spins forming AX-type spin systems that are foundational for DNP and quantum sensing.

Computation of Overhauser Dynamic Nuclear Polarization processes reveals fundamental correlation between water dynamics, structure, and solvent restructuring entropy
D. C. R. Brown, T. R. Webber, T. M. Casey, J. Franck, M. S. Shell, and S. G. Han Phys. Chem. Chem. Phys., 26 (20) (2024), 14637–14650.
https://doi.org/10.1039/D4CP00030G. PMID: 38742831.
Hydration water dynamics, structure, and thermodynamics are crucially important to understand and predict water-mediated properties at molecular interfaces. Yet experimentally and directly quantifying water behavior locally near interfaces at the sub-nanometer scale is challenging, especially at interfaces submerged in biological solutions. Overhauser dynamic nuclear polarization (ODNP) experiments measure equilibrium hydration water dynamics within 8–15 angstroms of a nitroxide spin probe on instantaneous timescales (10 picoseconds to nanoseconds), making ODNP a powerful tool for probing local water dynamics in the vicinity of the spin probe. As with other spectroscopic techniques, concurrent computational analysis is necessary to gain access to detailed molecular level information about the dynamic, structural, and thermodynamic properties of water from experimental ODNP data. We chose a model system that can systematically tune the dynamics of water, a water–glycerol mixture with compositions ranging from 0 to 0.3 mole fraction glycerol. We demonstrate the ability of molecular dynamics (MD) simulations to compute ODNP spectroscopic quantities, and show that translational, rotational, and hydrogen bonding dynamics of hydration water align strongly with spectroscopic ODNP parameters. Moreover, MD simulations show tight correlations between the dynamic properties of water that ODNP captures and the structural and thermodynamic behavior of water. Hence, experimental ODNP readouts of varying water dynamics suggest changes in local structural and thermodynamic hydration water properties.
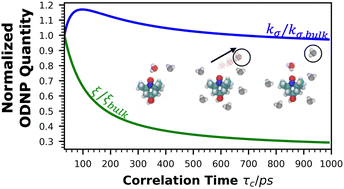
Computation of Overhauser Dynamic Nuclear Polarization processes reveals fundamental correlation between water dynamics, structure, and solvent restructuring entropy
D. C. R. Brown, T. R. Webber, T. M. Casey, J. Franck, M. S. Shell, and S. G. Han Phys. Chem. Chem. Phys., 26 (20) (2024), 14637–14650.
https://doi.org/10.1039/D4CP00030G. PMID: 38742831.
Hydration water dynamics, structure, and thermodynamics are crucially important to understand and predict water-mediated properties at molecular interfaces. Yet experimentally and directly quantifying water behavior locally near interfaces at the sub-nanometer scale is challenging, especially at interfaces submerged in biological solutions. Overhauser dynamic nuclear polarization (ODNP) experiments measure equilibrium hydration water dynamics within 8–15 angstroms of a nitroxide spin probe on instantaneous timescales (10 picoseconds to nanoseconds), making ODNP a powerful tool for probing local water dynamics in the vicinity of the spin probe. As with other spectroscopic techniques, concurrent computational analysis is necessary to gain access to detailed molecular level information about the dynamic, structural, and thermodynamic properties of water from experimental ODNP data. We chose a model system that can systematically tune the dynamics of water, a water–glycerol mixture with compositions ranging from 0 to 0.3 mole fraction glycerol. We demonstrate the ability of molecular dynamics (MD) simulations to compute ODNP spectroscopic quantities, and show that translational, rotational, and hydrogen bonding dynamics of hydration water align strongly with spectroscopic ODNP parameters. Moreover, MD simulations show tight correlations between the dynamic properties of water that ODNP captures and the structural and thermodynamic behavior of water. Hence, experimental ODNP readouts of varying water dynamics suggest changes in local structural and thermodynamic hydration water properties.

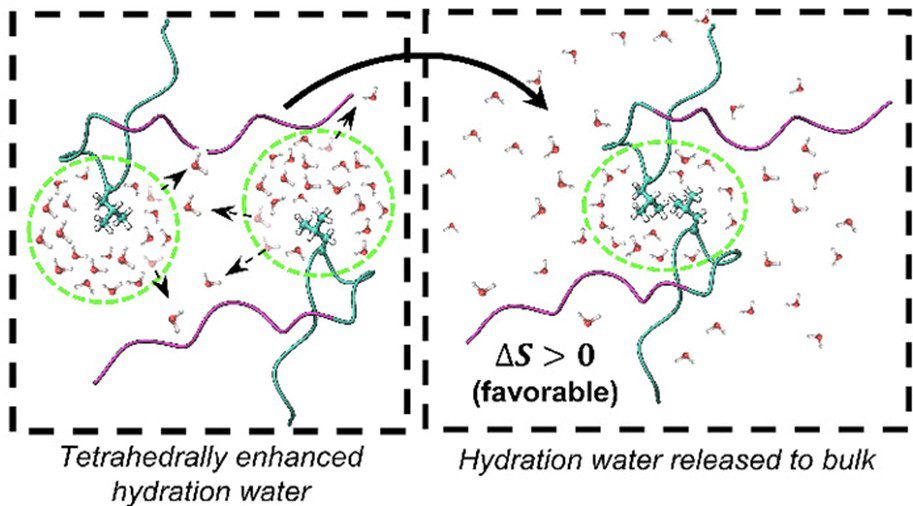
Bridging the Gap in Cryopreservation Mechanism: Unraveling the Interplay between Structure, Dynamics, and Thermodynamics in Cryoprotectant Aqueous Solutions
Cryoprotectants play a crucial role in preserving biological material, ensuring their viability during storage and facilitating crucial applications such as the conservation of medical compounds, tissues, and organs for transplantation. However, the precise mechanism by which cryoprotectants modulate the thermodynamic properties of water to impede the formation and growth of ice crystals, thus preventing long-term damage, remains elusive. This is evident in the use of empirically optimized recipes for mixtures that typically contain DMSO, glycerol, and various sugar constituents. Here, we use terahertz calorimetry, Overhauser nuclear polarization, and molecular dynamics simulations to show that DMSO exhibits a robust structuring effect on water around its methyl groups, reaching a maximum at a DMSO mole fraction of XDMSO = 0.33. In contrast, glycerol exerts a smaller water-structuring effect, even at higher concentrations (Scheme 1). These results potentially suggest that the wrapped water around DMSO’s methyl group, which can be evicted upon ligand binding, may render DMSO a more surface-active cryoprotectant than glycerol, while glycerol may participate more as a viscogen that acts on the entire sample. These findings shed light on the molecular intricacies of cryoprotectant solvation behavior and have potentially significant implications for optimizing cryopreservation protocols.
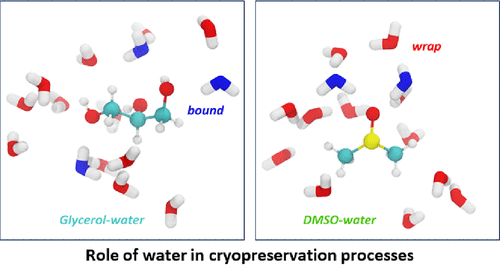
Bridging the Gap in Cryopreservation Mechanism: Unraveling the Interplay between Structure, Dynamics, and Thermodynamics in Cryoprotectant Aqueous Solutions
Cryoprotectants play a crucial role in preserving biological material, ensuring their viability during storage and facilitating crucial applications such as the conservation of medical compounds, tissues, and organs for transplantation. However, the precise mechanism by which cryoprotectants modulate the thermodynamic properties of water to impede the formation and growth of ice crystals, thus preventing long-term damage, remains elusive. This is evident in the use of empirically optimized recipes for mixtures that typically contain DMSO, glycerol, and various sugar constituents. Here, we use terahertz calorimetry, Overhauser nuclear polarization, and molecular dynamics simulations to show that DMSO exhibits a robust structuring effect on water around its methyl groups, reaching a maximum at a DMSO mole fraction of XDMSO = 0.33. In contrast, glycerol exerts a smaller water-structuring effect, even at higher concentrations (Scheme 1). These results potentially suggest that the wrapped water around DMSO’s methyl group, which can be evicted upon ligand binding, may render DMSO a more surface-active cryoprotectant than glycerol, while glycerol may participate more as a viscogen that acts on the entire sample. These findings shed light on the molecular intricacies of cryoprotectant solvation behavior and have potentially significant implications for optimizing cryopreservation protocols.

Nanoscale Water–Polymer Interactions Tune Macroscopic Diffusivity of Water in Aqueous Poly(Ethylene Oxide) Solutions
J. D. Moon, T. R. Webber, D. R. Brown, P. M. Richardson, T. M. Casey, R. A. Segalman, M. S. Shell, and S. G. Han Chem. Sci., 15 (7) (2024), 2495–2508.
https://doi.org/10.1039/D3SC05377F. PMID: 38362435. PMCID: PMC10866362.
The separation and anti-fouling performance of water purification membranes is governed by both macroscopic and molecular-scale water properties near polymer surfaces. However, even for poly(ethylene oxide) (PEO) – ubiquitously used in membrane materials – there is little understanding of whether or how the molecular structure of water near PEO surfaces affects macroscopic water diffusion. Here, we probe both time-averaged bulk and local water dynamics in dilute and concentrated PEO solutions using a unique combination of experimental and simulation tools. Pulsed-Field Gradient NMR and Overhauser Dynamic Nuclear Polarization (ODNP) capture water dynamics across micrometer length scales in sub-seconds to sub-nanometers in tens of picoseconds, respectively. We find that classical models, such as the Stokes-Einstein and Mackie-Meares relations, cannot capture water diffusion across a wide range of PEO concentrations, but that free volume theory can. Our study shows that PEO concentration affects macroscopic water diffusion by enhancing the water structure and altering free volume. ODNP experiments reveal that water diffusivity near PEO is slower than in the bulk in dilute solutions, previously not recognized by macroscopic transport measurements, but the two populations converge above the polymer overlap concentration. Molecular dynamics simulations reveal that the reduction in water diffusivity occurs with enhanced tetrahedral structuring near PEO. Broadly, we find that PEO does not simply behave like a physical obstruction but directly modifies water’s structural and dynamic properties. Thus, even in simple PEO solutions, molecular scale structuring and the impact of polymer interfaces is essential to capturing water diffusion, an observation with important implications for water transport through structurally complex membrane materials.
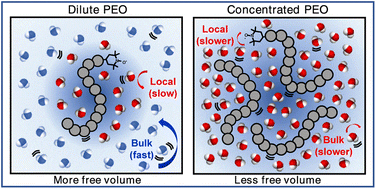
Nanoscale Water–Polymer Interactions Tune Macroscopic Diffusivity of Water in Aqueous Poly(Ethylene Oxide) Solutions
J. D. Moon, T. R. Webber, D. R. Brown, P. M. Richardson, T. M. Casey, R. A. Segalman, M. S. Shell, and S. G. Han Chem. Sci., 15 (7) (2024), 2495–2508.
https://doi.org/10.1039/D3SC05377F. PMID: 38362435. PMCID: PMC10866362.
The separation and anti-fouling performance of water purification membranes is governed by both macroscopic and molecular-scale water properties near polymer surfaces. However, even for poly(ethylene oxide) (PEO) – ubiquitously used in membrane materials – there is little understanding of whether or how the molecular structure of water near PEO surfaces affects macroscopic water diffusion. Here, we probe both time-averaged bulk and local water dynamics in dilute and concentrated PEO solutions using a unique combination of experimental and simulation tools. Pulsed-Field Gradient NMR and Overhauser Dynamic Nuclear Polarization (ODNP) capture water dynamics across micrometer length scales in sub-seconds to sub-nanometers in tens of picoseconds, respectively. We find that classical models, such as the Stokes-Einstein and Mackie-Meares relations, cannot capture water diffusion across a wide range of PEO concentrations, but that free volume theory can. Our study shows that PEO concentration affects macroscopic water diffusion by enhancing the water structure and altering free volume. ODNP experiments reveal that water diffusivity near PEO is slower than in the bulk in dilute solutions, previously not recognized by macroscopic transport measurements, but the two populations converge above the polymer overlap concentration. Molecular dynamics simulations reveal that the reduction in water diffusivity occurs with enhanced tetrahedral structuring near PEO. Broadly, we find that PEO does not simply behave like a physical obstruction but directly modifies water’s structural and dynamic properties. Thus, even in simple PEO solutions, molecular scale structuring and the impact of polymer interfaces is essential to capturing water diffusion, an observation with important implications for water transport through structurally complex membrane materials.

Precision Proteoform Design for 4R Tau Isoform Selective Templated Aggregation
Prion-like spread of disease-specific tau conformers is a hallmark of all tauopathies. A 19-residue probe peptide containing a P301L mutation and spanning the R2/R3 splice junction of tau folds and stacks into seeding-competent fibrils and induces aggregation of 4R, but not 3R tau. These tau peptide fibrils propagate aggregated intracellular tau over multiple generations, have a high β-sheet content, a colocalized lipid signal, and adopt a well-defined U-shaped fold found in 4R tauopathy brain-derived fibrils. Fully atomistic replica exchange molecular dynamics (MD) simulations were used to compute the free energy landscapes of the conformational ensemble of the peptide monomers. These identified an aggregation-prohibiting β-hairpin structure and an aggregation-competent U-fold unique to 4R tauopathy fibrils. Guided by MD simulations, we identified that the N-terminal-flanking residues to PHF6, which slightly vary between 4R and 3R isoforms, modulate seeding. Strikingly, when a single amino acid switch at position 305 replaced the serine of 4R tau with a lysine from the corresponding position in the first repeat of 3R tau, the seeding induced by the 19-residue peptide was markedly reduced. Conversely, a 4R tau mimic with three repeats, prepared by replacing those amino acids in the first repeat with those amino acids uniquely present in the second repeat, recovered aggregation when exposed to the 19-residue peptide. These peptide fibrils function as partial prions to recruit naive 4R tau—ten times the length of the peptide—and serve as a critical template for 4R tauopathy propagation. These results hint at opportunities for tau isoform–specific therapeutic interventions.
Precision Proteoform Design for 4R Tau Isoform Selective Templated Aggregation
Prion-like spread of disease-specific tau conformers is a hallmark of all tauopathies. A 19-residue probe peptide containing a P301L mutation and spanning the R2/R3 splice junction of tau folds and stacks into seeding-competent fibrils and induces aggregation of 4R, but not 3R tau. These tau peptide fibrils propagate aggregated intracellular tau over multiple generations, have a high β-sheet content, a colocalized lipid signal, and adopt a well-defined U-shaped fold found in 4R tauopathy brain-derived fibrils. Fully atomistic replica exchange molecular dynamics (MD) simulations were used to compute the free energy landscapes of the conformational ensemble of the peptide monomers. These identified an aggregation-prohibiting β-hairpin structure and an aggregation-competent U-fold unique to 4R tauopathy fibrils. Guided by MD simulations, we identified that the N-terminal-flanking residues to PHF6, which slightly vary between 4R and 3R isoforms, modulate seeding. Strikingly, when a single amino acid switch at position 305 replaced the serine of 4R tau with a lysine from the corresponding position in the first repeat of 3R tau, the seeding induced by the 19-residue peptide was markedly reduced. Conversely, a 4R tau mimic with three repeats, prepared by replacing those amino acids in the first repeat with those amino acids uniquely present in the second repeat, recovered aggregation when exposed to the 19-residue peptide. These peptide fibrils function as partial prions to recruit naive 4R tau—ten times the length of the peptide—and serve as a critical template for 4R tauopathy propagation. These results hint at opportunities for tau isoform–specific therapeutic interventions.
