Design spin cluster for NMR signal amplification and quantum resonance sensing
To reveal “invisible” NMR signal of surfaces, active sites, and functional species in catalysis, molecular recognition and quantum materials using out of the box tools.
The Han lab pushes the frontier of magnetic resonance to discover new chemistry principles in water and through control over the shaping and ordering of dynamic water. We use advanced magnetic resonance manipulation and control over the spatial organization of electron and nuclear spin clusters located on biomolecular surface, soft materials or nanomaterials to uncover their structure, the design rules for molecular recognition, and the surface structuring and dynamics of hydration water.
The development effort requires multiple research tools in the realm of physical chemistry broadly speaking. They include instrument development to achieve hyperpolarization and quantum resonance sensing, the design of precisely tuned electron and nuclear spin clusters, spin physic theory and simulations, and the dynamics and thermodynamics of solvation science to control biomolecular activity and directed assembly.
The Han lab is pushing the frontier of electron and nuclear spin magnetic resonance instrumentation and concepts in dynamic nuclear polarization (DNP) amplified nuclear magnetic resonance (NMR) and electron paramagnetic resonance (EPR). We are motivated by the power of “Seeing is Believing”. Visualizing molecular interactions and materials interfaces, previously “invisible”, can fundamentally transform our ability to discover solutions, and almost as importantly, ask new questions.

Research in the Han Lab builds and employs state-of-the-art tools in magnetic resonance spectroscopy to advance our understanding in different subject areas, ranging from quantum sensing, solvation science, biophysics to neurodegenerative diseases.
SEE ALL RESEARCHTo reveal “invisible” NMR signal of surfaces, active sites, and functional species in catalysis, molecular recognition and quantum materials using out of the box tools.
To understand, control and engineer protein aggregation pathways, protein surface activity to protein liquid-liquid phase separation.
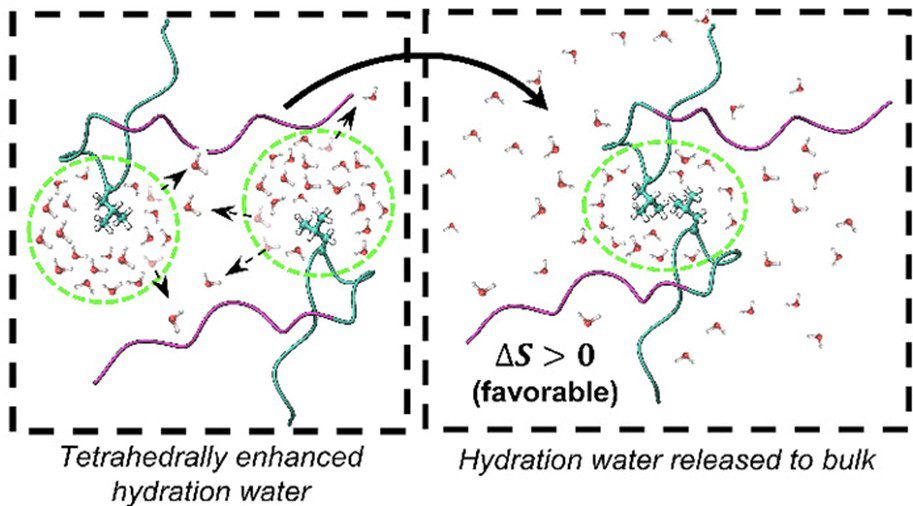
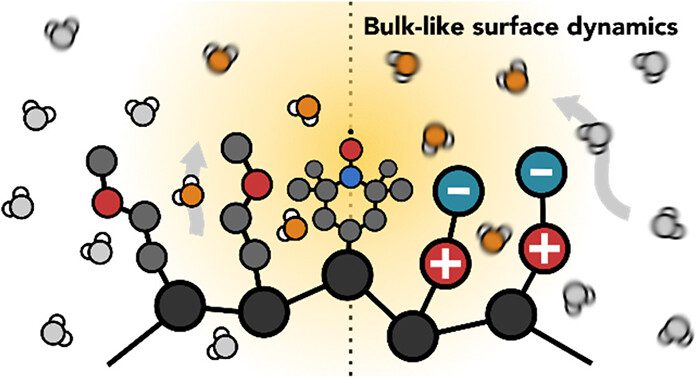
To reveal long-standing questions on the structure and dynamics of water on proteins, membranes to catalyst support surfaces.
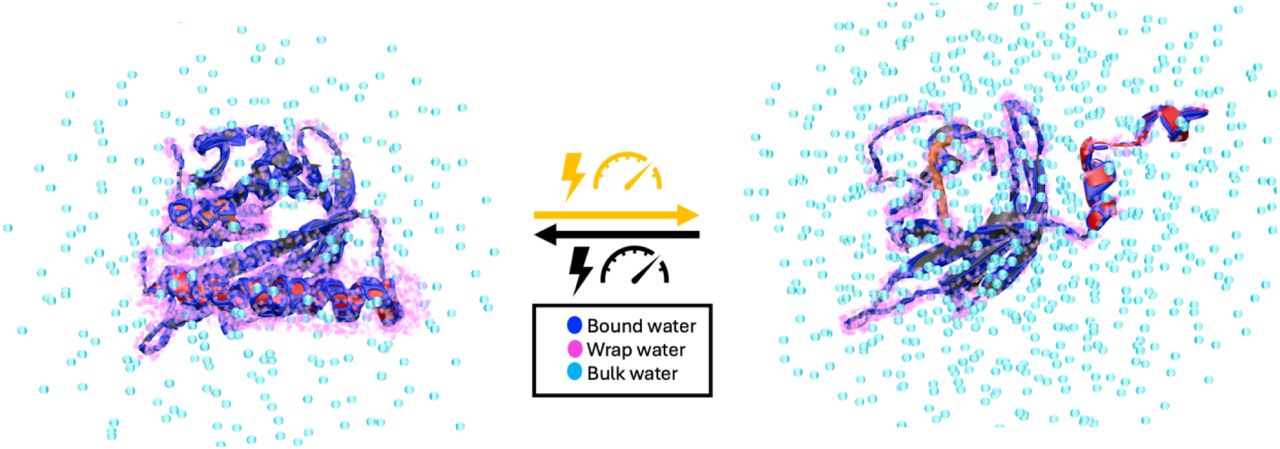
Tau forms fibrillar aggregates that are pathological hallmarks of a family of neurodegenerative diseases known as tauopathies. The synthetic replication of disease-specific fibril structures is a critical gap for developing diagnostic and therapeutic tools. This study debuts a strategy of identifying a critical and minimal folding motif in fibrils characteristic of tauopathies and generating seeding-competent fibrils from the isolated tau peptides. The 19-residue jR2R3 peptide (295 to 313) which spans the R2/R3 splice junction of tau, and includes the P301L mutation, is one such peptide that forms prion-competent fibrils. This tau fragment contains the hydrophobic VQIVYK hexapeptide that is part of the core of all known pathological tau fibril structures and an intramolecular counterstrand that stabilizes the strand–loop–strand (SLS) motif observed in 4R tauopathy fibrils. This study shows that P301L exhibits a duality of effects: it lowers the barrier for the peptide to adopt aggregation-prone conformations and enhances the local structuring of water around the mutation site to facilitate site-directed pinning and dewetting around sites 300-301 to achieve in-register stacking of tau to cross β-sheets. We solved a 3 Å cryo-EM structure of jR2R3-P301L fibrils in which each protofilament layer contains two jR2R3-P301L copies, of which one adopts a SLS fold found in 4R tauopathies and the other wraps around the SLS fold to stabilize it, reminiscent of the three- and fourfold structures observed in 4R tauopathies. These jR2R3-P301L fibrils are competent to template full-length 4R tau in a prion-like manner.
Harvesting water from air offers a promising solution to the global water crisis. However, existing sorbents often struggle in arid climates due to limitations such as low sorption capacities, hydrolytic instability, slow mass transport, high desorption enthalpy, and costly operation. Phosphonate-based metal–organic frameworks (MOFs), known for their exceptional water stability, have not been extensively explored for water harvesting. This study systematically investigates the performance of STA-12 (M═Co, Ni, Mg) and STA-16 (M═Co, Ni), a series of stable phosphonate-based MOFs, as water sorbents. STA-12 MOFs demonstrate remarkable adsorption at ultra-low humidity (<10%), while STA-16(Co) exhibits a high water uptake capacity of 0.54 g g−1 at 10–50% relative humidity (RH) and 0.72 g g−1 at 34% RH. Molecular simulations and solid-state NMR identified liquid-like water, critical for harvesting applications, as the key contributor to the superior sorption performance of STA-16(Co). Scalable aqueous synthesis methods are developed, producing tens of grams of MOFs per batch without high-pressure equipment. A prototype device incorporating STA-12(Ni) demonstrated the feasibility of these materials for real-world water harvesting, showcasing their potential to address water scarcity in arid regions