Quantum Control and Sensing by Spin Cooling
To reveal “invisible” NMR signal of surfaces, active sites, and functional species in catalysis, molecular recognition and quantum materials using out of the box tools.
The Han Lab pushes the frontiers of magnetic resonance and quantum information science with the goal of “seeing” chemical and biological building blocks and processes at the quantum limit. The two main themes of the Han Lab over the past 20 years have been spins and water. Electron and nuclear spins are the ultimate quantum reporters and contrast agents for biochemical processes and chemical building blocks. Advanced magnetic resonance sensing, control over the spatial organization of electron and nuclear spin clusters, and dual electron-nuclear magnetic resonance techniques have contributed to uncovering new design rules for molecular recognition, as well as the surface structuring, shaping, and ordering of biological water.
Recently, we have begun to ask the ultimate question: do quantum phenomena direct and control biological and chemical processes? The answer is yes, but high-quality experimental validations are key to qualifying the context and boundaries of these answers. Recent breakthrough developments by the Han Lab offer one-of-a-kind experimental tools that allow us to gain control over the initialization and manipulation of quantum spin states via spin cooling at high magnetic fields.
This development effort requires interdisciplinary research tools, including instrument development to combine electron and nuclear magnetic resonance with optical excitation and detection, the design of precisely tuned electron and nuclear spin qubits, spin dynamics simulations, and studies of the dynamics and thermodynamics of solvation to control biomolecular activity and assembly.
We are motivated by the power of “Seeing is Believing.” New tools for visualizing molecular interactions and materials interfaces—previously “invisible”—have fundamentally transformed our ability to discover solutions and ask new questions. The next frontier of visualization will be quantum microscopy that relies on spins as quantum reporters to deliver molecular insights that conventional microscopy cannot.

Research in the Han Lab develops novel tools to exploit spins as quantum reporters with unprecedented sensitivity and information content and as biological qubits with spin state control. Our core interest lies in advancing spin-based quantum information science, solvation science, and the molecular basis of signal transduction.
SEE ALL RESEARCHTo reveal “invisible” NMR signal of surfaces, active sites, and functional species in catalysis, molecular recognition and quantum materials using out of the box tools.
To reveal long-standing questions on the structure and dynamics of water on proteins, membranes to catalyst support surfaces.
To understand, control and engineer protein aggregation pathways, protein surface activity to protein liquid-liquid phase separation.
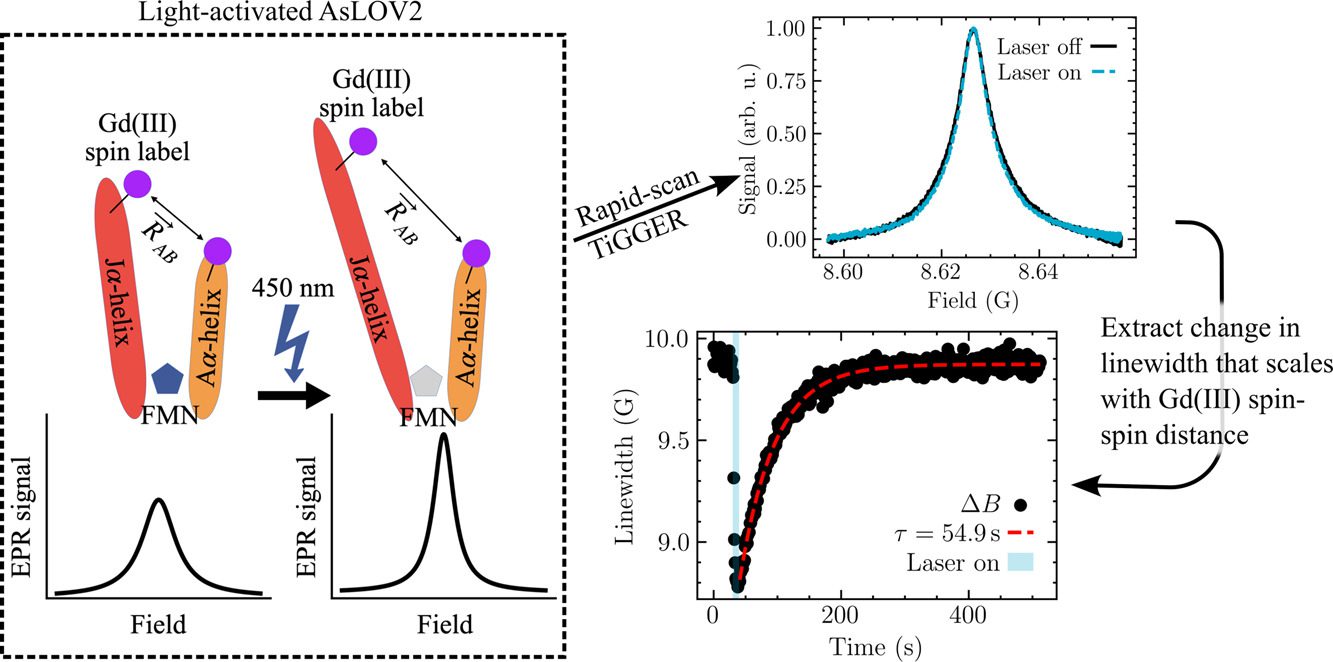
We present field-domain rapid-scan (RS) electron paramagnetic resonance (EPR) at 8.6 T and 240 GHz. To enable this technique, we upgraded a homebuilt EPR spectrometer with an FPGA-enabled digitizer and real-time processing software. The software leverages the Hilbert transform to recover the in-phase (𝐼) and quadrature (𝑄) channels, and therefore the raw absorptive and dispersive signals, 𝜒′ and 𝜒′′, from their combined magnitude (√𝐼2+𝑄2). Averaging a magnitude is simpler than real-time coherent averaging and has the added benefit of permitting long-timescale signal averaging (up to at least 2.5 × 106 scans) because it eliminates the effects of sourcereceiver phase drift. Our rapid-scan (RS) EPR provides a signal-to-noise ratio that is approximately twice that of continuous wave (CW) EPR under the same experimental conditions, after scaling by the square root of acquisition time. We apply our RS EPR as an extension of the recently reported time-resolved Gd-Gd EPR (TiGGER) [Maity et al., 2023], which is able to monitor inter-residue distance changes during the photocycle of a photoresponsive protein through changes in the Gd-Gd dipolar couplings. RS, opposed to CW, returns field-swept spectra as a function of time with 10 ms time resolution, and thus, adds a second dimension to the static field transients recorded by TiGGER. We were able to use RS TiGGER to track time-dependent and temperature-dependent kinetics of AsLOV2, a light-activated phototropin domain found in oats. The results presented here combine the benefits of RS EPR with the improved spectral resolution and sensitivity of Gd chelates at high magnetic fields. In the future, field-domain RS EPR at high magnetic fields may enable studies of other real-time kinetic processes with time resolutions that are otherwise difficult to access in the solution state.
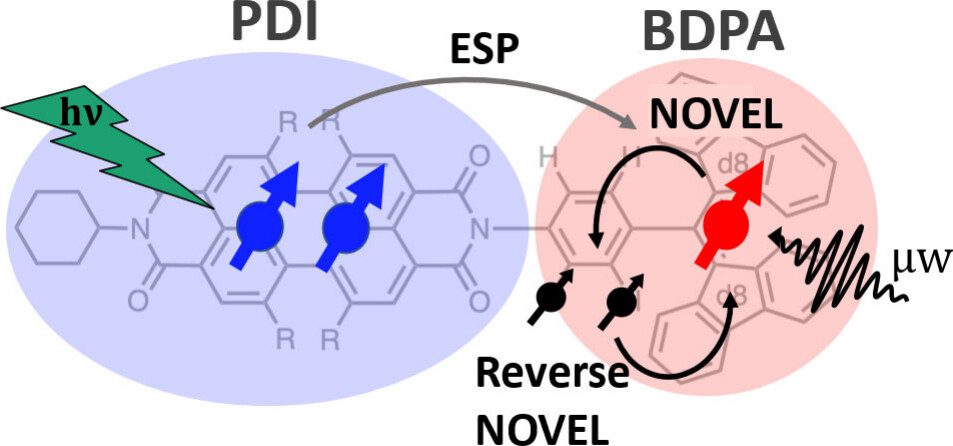
Chromophore radicals (CR) are emerging as important components for molecular quantum information science (QIS), especially in the context of quantum sensing. Here, we demonstrate that the optically hyperpolarized electrons in a 1,6,7,12-tetrakis(4-tert-butylphenoxy)-perylene-3,4,9,10-bis(dicarboximide) (tpPDI) covalently linked to a partially deuterated 1,3-bis(diphenylene)-d16-2-phenylallyl radical (BDPA-d16) can be coherently manipulated via pulsed dynamic nuclear polarization (DNP) methods to transfer polarization to nuclear spins and back. Under light illumination at 85 K, electron hyperpolarization in BDPA is enhanced 2.1- to 2.4-fold over thermal polarization and lasts for more than 100 ms. By applying nuclear orientation via electron spin-locking (NOVEL) DNP, this optically amplified electron hyperpolarization was successfully transferred to a 1H nuclear spin within the CR system and efficiently returned to the electron spin for readout via reverse-NOVEL. The NOVEL transfer efficiency of 65% amounts to a 688-fold nuclear spin hyperpolarization of the target nuclear spin, considering the 2.1-fold electron spin hyperpolarization. This reversible coherent manipulation of hyperpolarization transfer highlights the utility of CR systems to initialize and read out nuclear spin states in a disordered matrix at moderate cryogenic temperatures. Coupled with CRs’ environmental compatibility, tunability, and precise state initialization, these results highlight the promising role of nuclear spins in CRs for QIS applications, including quantum sensing and memory.
A critical discovery of the past decade is that tau protein fibrils adopt disease-specific hallmark structures in each tauopathy. The faithful generation of synthetic fibrils adopting hallmark structures that can serve as targets for developing diagnostic and/or therapeutic strategies remains a grand challenge. We report on a rational design of synthetic fibrils built of a short peptide that adopts a critical structural motif in tauopathy fibrils found in Alzheimer’s Disease (AD) and Chronic Traumatic Encephalopathy (CTE). They serve as minimal prions with exquisite seeding competency, in vitro and in tau biosensor cells, for recruiting tau constructs ten times larger its size en route to AD or CTE fibril structures. We demonstrate that the generation of AD and CTE-like fibril structures is dramatically catalyzed in the presence of mini-AD prions and further influenced by salt composition in solution. Double Electron-Electron Resonance studies confirmed the preservation of AD-like folds across multi-generational seeding. Fibrils formed with the full AD/CTE-like core show strong seeding competency, with their templating effect dominating over the choice of salt composition that tunes the initial selection of AD- and CTE-like fibril populations. The mini-AD prions serve as a potent catalyst with templating capabilities that offer a novel strategy to design pathological tau fibril models.